Imaris Open
FILE EXCHANGE & FORUM
Frequently Asked Questions
Imaris 8.3 includes a set of key tools, improved for almost 25 years, which cater for the needs of researchers in live cell imaging and, in particular, developmental biology.
Using the Imaris Cell module researchers have been able to automatically identify cells based on cytoplasmic, nuclear and membrane labels. Up until recently the membrane based detection required a nuclear marker to automatically segment each individual cell. In Imaris 8.3 we introduced a new membrane detection that detect cells faster and without the need for a nuclear marker. We have also introduced new modification options to split/merge detected cells as well as Object ID color coding and a new visualization option - slicer display.
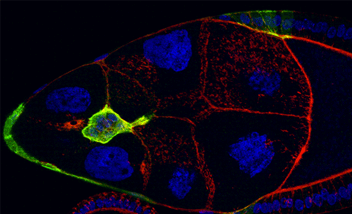
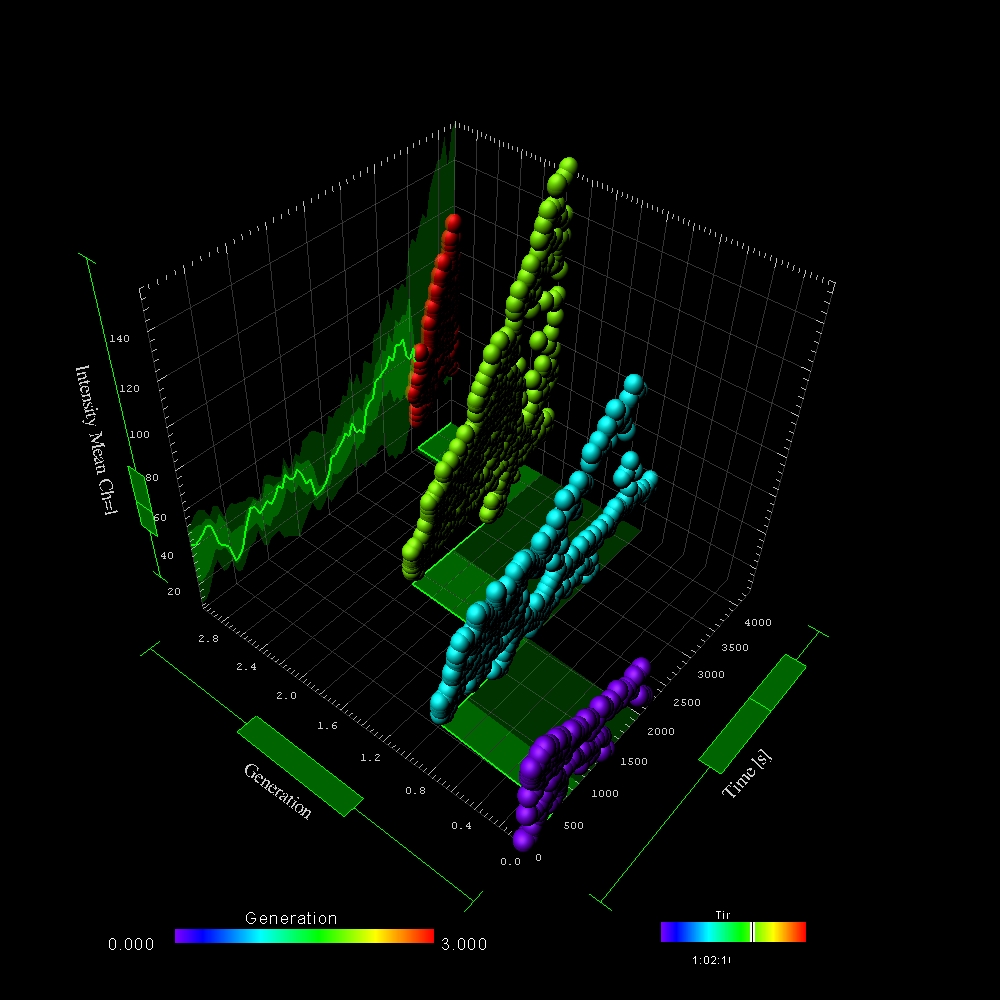
Imaris MeasurementPro enables researchers to extract critical statistical parameters from their microscopy images thus allowing for the quantification of scientific findings. Imaris MeasurementPro adds shape, size, and intensity based measurement capabilities to the volume rendering, surface rendering and object detection features of Imaris. It allows researchers to interactively classify, group, and filter segmented objects based on any of the calculated statistics. MeasurementPro enables 3D distance and angle measurements in your dataset.
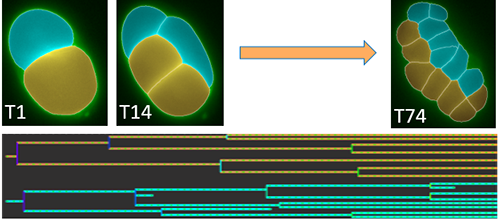
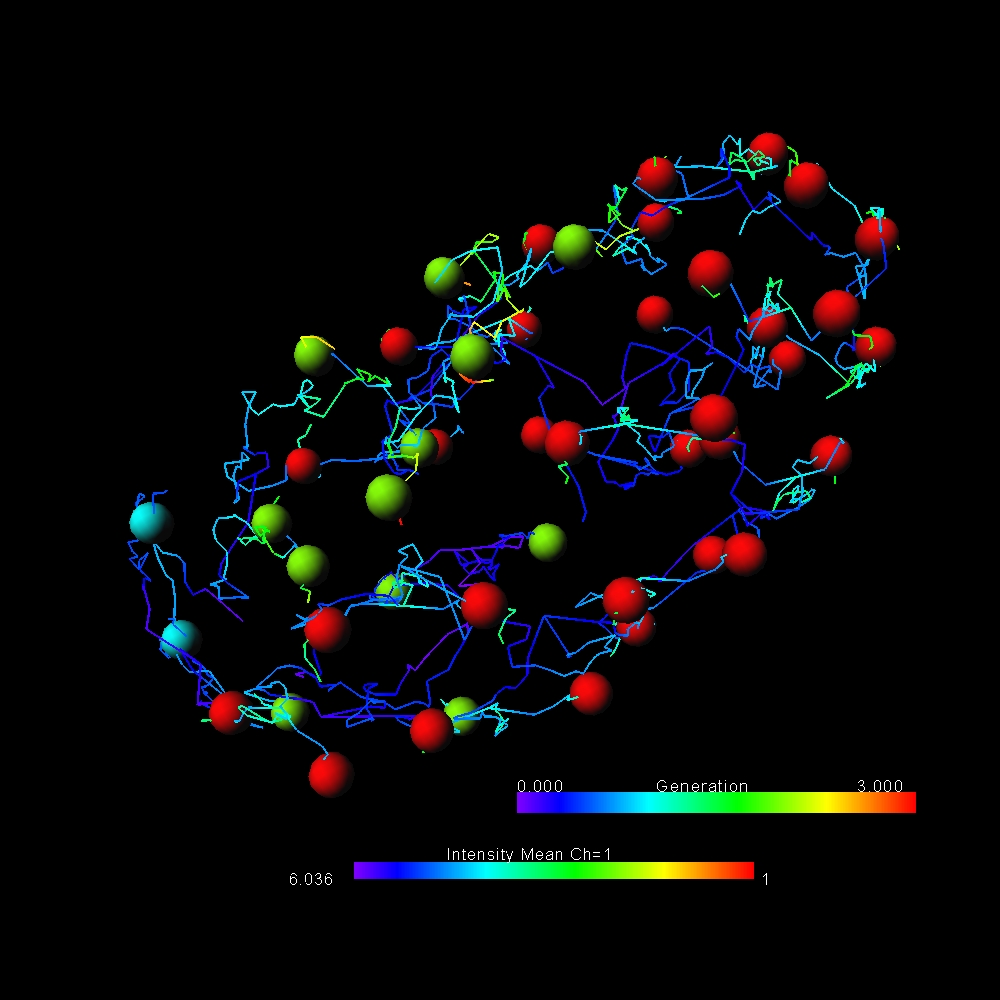
Imaris 8.3 includes a new Reference Frame feature providing users with a simple way to calculate distance/proximity based on a user specified coordinate system. The reference frame can be set using translational and rotational adjustment. Using this new features distance/proximity of structures of interest can automatically calculated based on your reference frame.
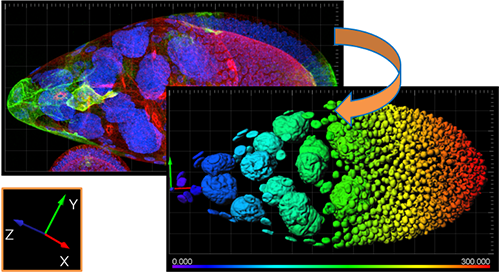
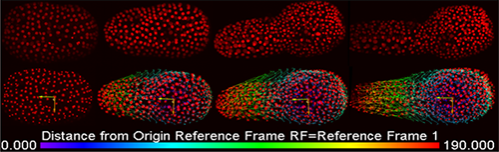
The new reference frame feature can also be used to re-align your data. Whether it is a single timepoint or a time series, reference frames can be placed manually or automatically and adjusted through time for alignment purposes. Tracking statistics can also be calculated based on the reference frame coordinate axis to correct for movement in the sample or to measure the relationship with a plane, or point, of interest. Providing accurate speed, distance and displacement statistics.
For those working with big data we have vastly improved the speed of conversion of your multi-dimensional datasets into the Imaris file format (.ims). Using the new Imaris File Converter application, converting into .ims will take a fraction of the time. This will make file opening, visualization and processing of large files more manageable. If you are interested in learning more about the latest Imaris release or would like a demonstration of the new features please let me know. Also if you would like to receive a maintenance renewal quote so you can up to date with all the latest Imaris features please click on the link below. If you are interested in learning more about the latest Imaris release, would like a demonstration of the new features or would like to receive a maintenance renewal quote so you can up to date with all the latest Imaris features please click on the green button below.
Discover our online collection of “How to...“ videos and and webinars presenting various features of Imaris that are useful for your data analysis.
Browse Imaris 8.3 XTension packs dedicated for cell and developmental biologists and download them free from Imaris Open to expand image analysis capabilities of your software.

XTensions included:

XTensions included:

XTensions included:
Learn more about interesting research in the field of developmental biology.



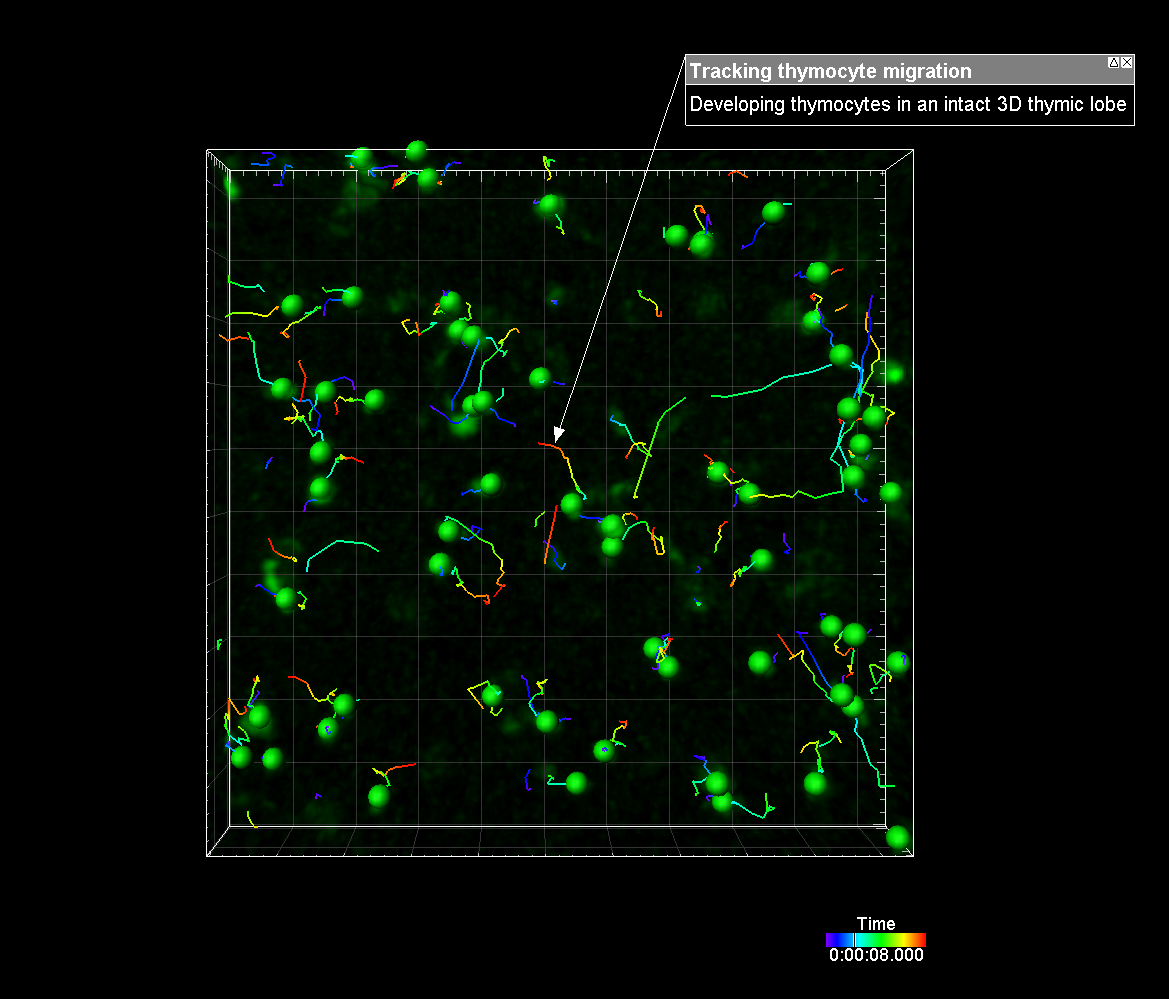
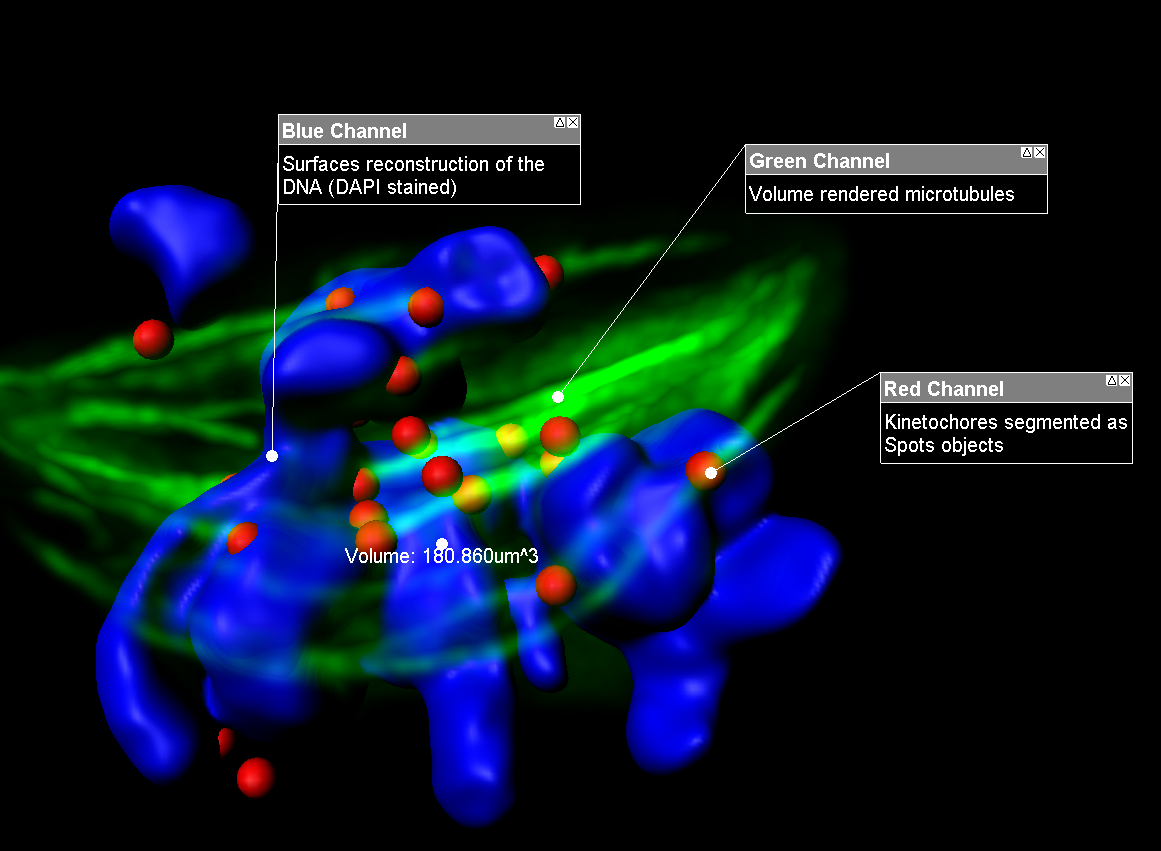
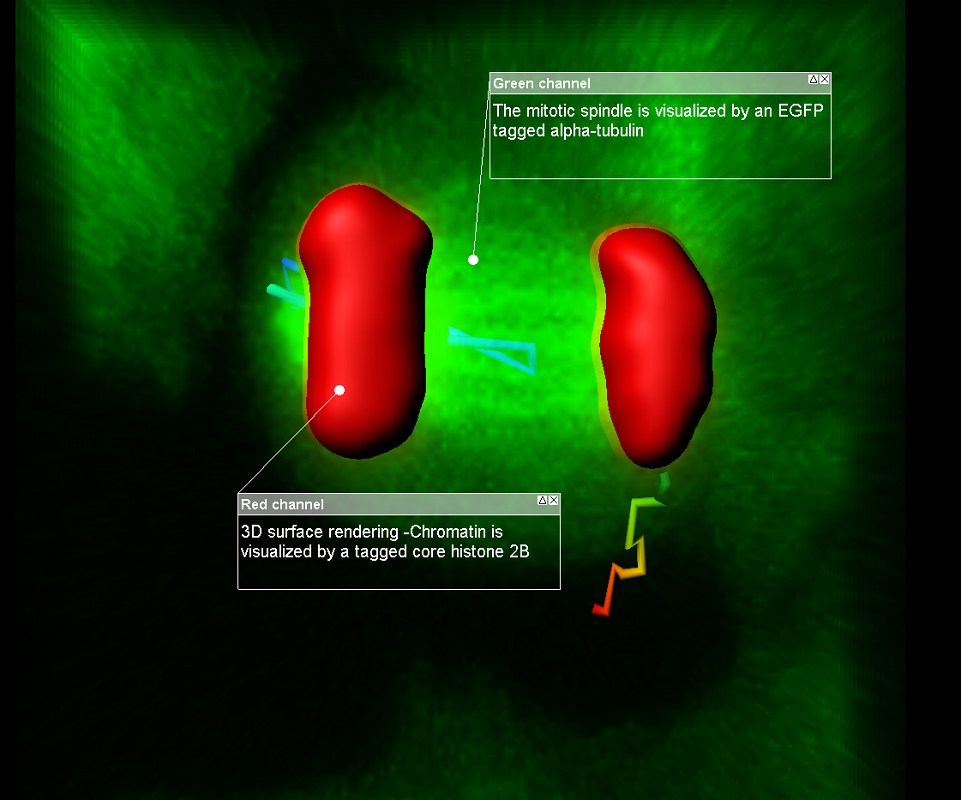
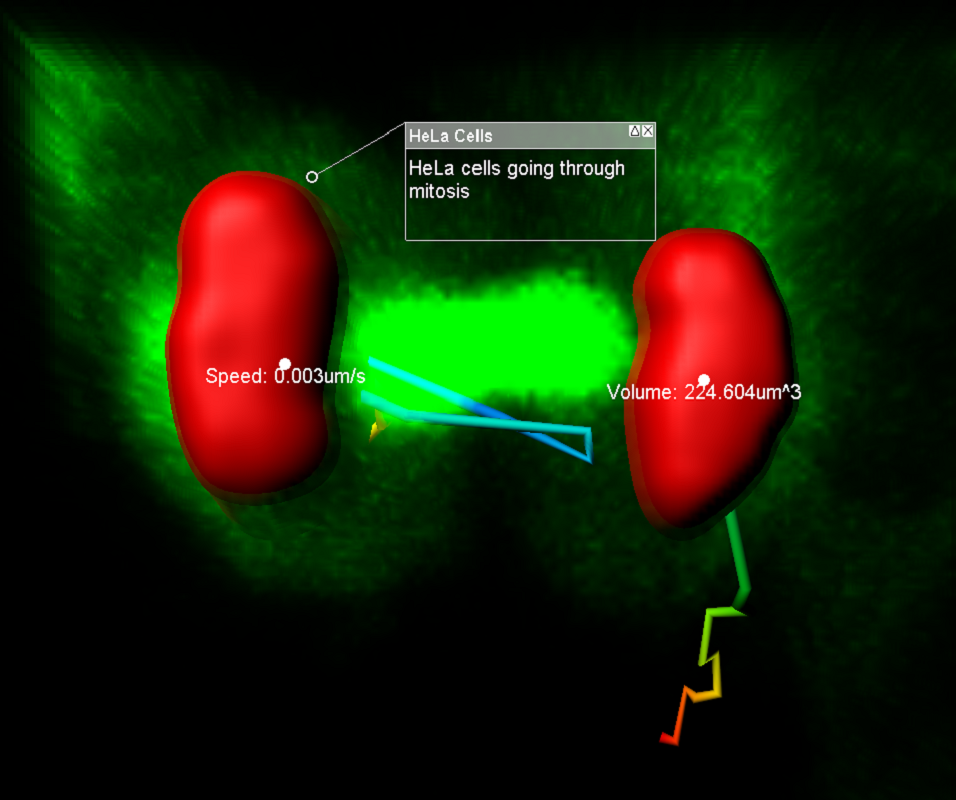
Join one of our local or online lectures by the top scientists presenting their recent research in the cell and developmental biology field. Meet Bitplane application specialists and ask questions specific to your images (onsite) or learn more about the new Imaris 8.3 features (online).